Guest post by Kathryn “Kaylee” Mueller, Phase Genomics.
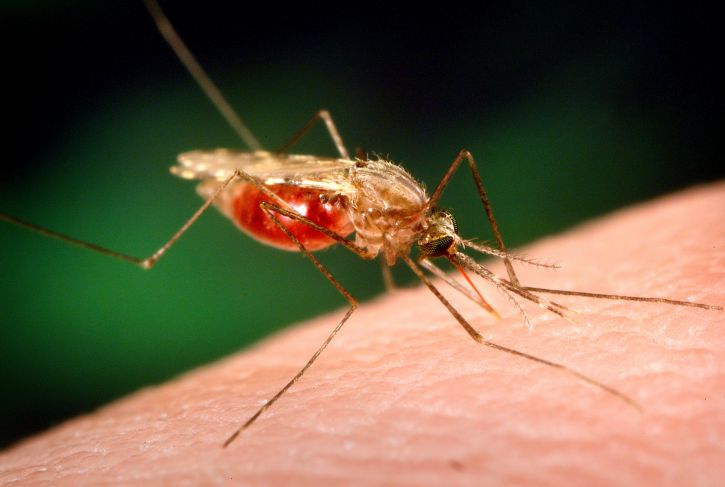
Plasmodium parasites—the microbes that cause malaria—are right at home in the tropics. After all, tropical regions harbor the two animals that the malaria parasites need to complete their complex lifecycle: female Anopheles mosquitoes and human beings. And in 2017 alone, Plasmodiumracked up 219 million cases of malaria, with 435,000 deaths.
Though malaria has spread over time across the world’s tropical areas, one region in particular—sub-Saharan Africa—unquestionably remains ground zero for the disease. African countries accounted for more than 90 percent of malaria cases and malaria deaths in 2017, according to the World Health Organization. Plasmodium‘s options for mosquito hosts in sub-Saharan Africa may explain malaria’s outsized presence there.
“Africa suffers disproportionately,” says Nora Besansky, a malaria researcher at the University of Notre Dame. “This is mainly due to the dominance of two highly efficient mosquito vectors of human malaria found throughout that region: Anopheles gambiae and Anopheles funestus.”
The twin scourges of malaria in Africa
Researchers long paid attention to An. gambiae due to its knack for transmitting the deadliest of the Plasmodium species. But scientists were slower to recognize that An. funestus, the mosquito that shares much of An. gambiae‘s range, also shows grim aptitude as a malaria vector. For example, An. funestus strongly prefers to bite humans over other animals. In addition, it tends to hang out indoors, gaining more opportunities to bite people and potentially spread malaria.
A game of catch-up
Yet scientists know much less about An. funestus than An. gambiae. Laboratory strains of An. funestus have been around for just 19 years, compared to more than 65 years for An. gambiae. The An. gambiae genome was released in 2002, the second for insects. The An. funestus genome came out in 2015, but its fragmented assembly consists of almost 1,400 scaffolds—despite a nuclear genome of just three chromosome pairs—resulting in fragmented genes, missing gene clusters, and little reliable structural information about the genome.
Assembling a better genome
Besansky is part of a team that decided to start over. They have produced a new, chromosome-scale assembly of the An. funestus genome—all from a pool of individuals of an An. funestus laboratory strain. The team used PacBio sequencing to produce long-read sequences, which they assembled into larger contigs and then filtered to remove duplicated sequences.
“Since this was a colony of mosquitos, there was high sequence duplication present in the data,” said Jay Ghurye, a member of the team and doctoral student at the University of Maryland, College Park. “We had to remove the duplications before scaffolding them using Hi-C data.”
That Hi-C data came from Phase Genomics, a partner in this project. Phase Genomics used its Proximo Hi-C approach to collapse the team’s contigs into longer scaffolds and—ultimately—produce three chromosome-length assemblies for An. funestus.
A complete roadmap
Based on contig and scaffold length alone, the new genome for An. funestusis a 100-fold improvement on the prior assembly, according to the researchers.
“Just as driving from New York to Los Angeles is facilitated by a roadmap, genetic research also is more powerful, efficient, and accurate if the 260 million puzzle pieces of nucleotides in the nuclear genome are properly ordered and oriented,” said Dr. Besansky.
The team hopes that this more complete picture of the An. funestus genome will help scientists understand what makes it such an effective malaria vector and come up with measures to countermand it.
One large to-do list for one little bug
Researchers already have a bevy of burning questions about An. funestus.
For instance, according to the World Health Organization, An. funestus is fast evolving resistance to the pyrethroid insecticides that are used to treat bed nets. The new genome includes features left out of the first assembly, such as genes arranged in tandem clusters. And at least one type of gene often found in these types of clusters—cytochrome P450 genes—has already been implicated in pyrethroid resistance in An. funestus.
Resistance is spreading in other ways too. Some An. funestus populations may have evolved behavioral adaptations to evade anti-mosquito controls. Scientists can now employ the new genome to help locate the genetic underpinnings of these traits.
With the new genome, researchers should also have an easier time detecting chromosomal rearrangements, such as inversions, among An. funestus populations.
“Inversions affect the spread of malaria, indirectly if not directly, because they typically contain hundreds or thousands of genes involved in climatic or local adaptation—allowing their mosquito carriers to fully exploit heterogeneous and otherwise challenging environments,” says Besansky.
An. funestus has obviously been busy, employing multiple approaches to dodge anti-mosquito measures. But all of these adaptations originate from the same source: the An. funestus genome. With that toolbox now completely sequenced, aligned, assembled and annotated, scientists may, at last, have the right blueprints to bring this malaria vector to heel.