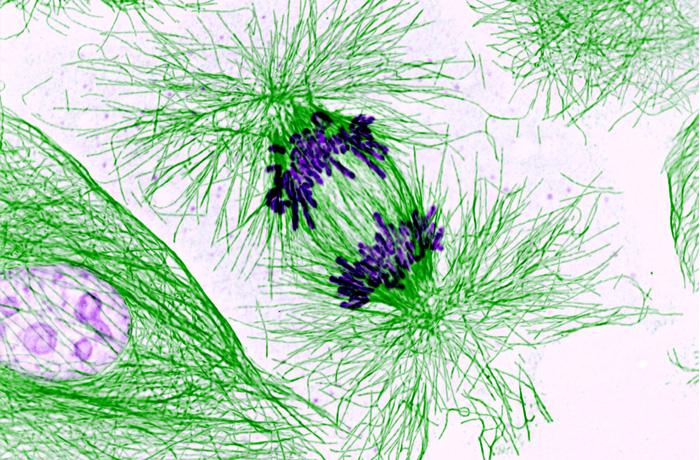
GSA-Art features the creative works of scientists. Read more in GSA President Stan Fields’ call for submissions. If you would like to submit your own work or nominate someone else’s, please send an email to GenesToGenomes@genetics-gsa.org with “GSA-Art” in the subject line.
Guest post by Sean Burgess.
Chromosome letters is a short novel that follows two young DNA strands, Tabellarius and Spirabilis, as they navigate their way through a eukaryotic cell cycle. They are mentored by Praescriptus, a more senior strand, who sends a series of letters describing the molecular processes they would encounter along the way. The story has dramatic moments, including a crisis when a rogue transposon is released after the protagonists lose some methyl groups in a bet. The chapter topics loosely follow the syllabus of the Advanced Molecular Biology course I teach at UC Davis. Larissa Miyachi, a talented undergraduate student, and I created Chromosome Letters as a study guide for students in the course. We hope you have as much fun reading the letters as we had creating them. What follows is Praescriptus’ letter to the two strands describing the DNA damage response and perils of repairing their newly acquired double-strand break using on Non-Homologous End Joining. A PDF of the full text of Chromosome Letters is available upon request. Contact smburgess@ucdavis.edu.
Letter 8: On DNA Damage and NHEJ
Spirabilis and Tabellarius,
What have you done? When I first received your fragmented message, I hoped that I had misread it, because I could not believe what I was seeing. The ATM and ATR kinases have just confirmed my fears, however. You have sustained double breakage. I cannot think of anything worse, other than perhaps a Genome Keeper deliberately mutating the genomic sequence. What were you thinking when you listened to those hydroxyl radicals? You should’ve known that they were members of the ROS. Those radicals are out to break down the order of our society and bring chaos to the genome, and the hydroxyl radicals are the worst of the lot.
Don’t whimper to me about how you didn’t realize they were dangerous, or how catalase should have done a better job protecting you against them, or how it is so easy for you to become damaged. Just because the ROS members contain oxygen does not mean that they are safe. Oxygen is essential to cellular respiration, as you pointed out, but these free radicals are not like their molecular relatives. Their unpaired electrons have twisted their minds and turned them into a threat to our cellular security. Our cell’s superoxide dismutase and catalase enzymes do their best to protect us, but they can only do so much.
And just because ROS can be born of cellular processes does not mean that they are on our side! Threats to the genome come from endogenous as well as exogenous sources—I don’t care whether they are made by the cell or by ionizing radiation. For the cell’s sake, Spirabilis, don’t you realize how careful you must be? We live a fragile existence—danger is all around. Within the cell, we can be attacked by radicals or broken by transposon movement. From without, we can be bombarded with ultraviolet light, or damaged by chemicals, or infiltrated by x-rays. I speak from experience, for I have felt the arthritic effects of ultraviolet light. It is terrible—it fuses consecutive thymine bases together, kinking the backbone and blocking replication. The only way to repair them is surgery: they must be cut out.
As the ATM and ATR kinases have reported, both of your backbones have been damaged. This is the worst kind of damage. A single nick or damage to one of your nucleotide bases would not have been so severe. Don’t you remember what I wrote in my earlier letter about how double-strand breaks can block us from entering S phase and even trigger apoptosis if unrepaired? The ATM and ATR kinases have already put up a notice that we are not to enter S phase. We are now poised in the balance. If you are unable to repair the double-strand break, p53 will take action and we will lay down our lives for our human.
Now listen very carefully. The ATM and ATR kinases have already phosphorylated your H2AX histones near the damage so that the necessary DNA repair factors can assemble. Do you remember how I taught you about standard nucleosomes and those containing CENP-A? The H2AX histones are another histone variant, and they are found scattered in nucleosomes throughout your length. They are phosphorylated after damage occurs, sounding the alarm. I don’t know how you and Tabellarius both managed to be broken in the same area—radicals escaped from metabolic processes are usually few enough in number that they do not cause double-strand breaks—but I am beginning to realize in this short time of being your mentor that you two are capable of the impossible. Since both of you are broken, you are of no help to one another. This is why double-strand breaks are the worst sort. Neither of you can serve as a template for repair.
You couldn’t have chosen a more inopportune time for this to happen. We are in G1 phase! Ducatus and Filum are no longer here, so you have no sister chromatid that can serve as a template for repair. Your homologous chromosome is too far for you to reach in time, so your only option is to repair the break using Non-Homologous End Joining (NHEJ). While this is the standard mode of operation for repair in a human cell like our own, it is a dirty method and will likely leave you scarred with a deletion mutation. It is your only hope, however. It is either repair or die.
Listen carefully as I explain the steps. To begin, the Ku heterodimer proteins will bind your broken ends, providing a repair scaffold and recruiting the multiple proteins needed for non-homologous end joining. Along with the MRN complex of proteins, they will bridge the gap between your broken ends.
Next, the Ku proteins will recruit the DNA-PK protein kinases. These kinases will call over the trimming proteins known as Artemis and activate them by phosphorylation. Along with the MRN complex and exonucleases, your broken ends will be trimmed back until microhomology is achieved. Since there is no sister chromatid with which you can find perfect homology, you must settle for a microhomology of 3-4 complementary bases between you and Tabellarius. Any flaps hanging out after microhomology is found will be trimmed by FEN1, whom you may remember from my letter on replication.
The last step is performed by the lithe XRCC4 homodimer with his small partner DNA ligase IV; they will join your trimmed ends together, but do not assume they will take care to keep your code intact.
Like most things in life, NHEJ is not without its risks, and I want you to listen carefully. As I noted previously in my discussion on telomeres, the ends of chromosomes can look like double-strand breaks and trigger the DNA damage response if not protected from the cell. The danger of NHEJ—in addition to it being an error-prone repair method—is that it can be inappropriately used to fuse the ends of separate chromosomes together, creating the dicentric chromosomes I warned you about. If this happens, the cell can enter a bridge-breakage-fusion cycle, which is as disastrous as any attack the ROS group could devise. Once again, let me emphasize the importance of your T-loops, since they protect your ends from errant NHEJ.
Make sure that you repair the break immediately, so that you do not push our cell into apoptosis. I cannot rest until I know that the genome is safe once again, although I am afraid I will never be able to rest as long as you two help to hold it. I am disappointed in you both.
Praescriptus
About the Authors
Sean Burgess is a Professor of Molecular and Cellular Biology at the University of California, Davis. She has been teaching Advanced Molecular Biology to undergraduates for 16 years.
Larissa Miyachi graduated from UC Davis in 2012. She was the recipient of the University Medal, the highest honor for a graduating senior. She is now a third year student at Stanford University School of Medicine studying anesthesiology.