For the past several decades, Shetland ponies’ collective past had caught up with them. A portion of the population of these miniature horses is affected by atavism, a phenomenon in which ancient characteristics are accidentally revived by mutations. Traits reincarnated in this way sometimes interact disastrously with the genetic background of the modern organism. For the ponies, a type of skeletal atavism (SA) has tragic consequences: the affected animals are born with some of the bones in their legs fused together, resulting in leg deformity and difficulty moving. Over ten percent of the ponies are carriers, and most affected individuals have to be euthanized at a young age. In an article in the July issue of G3, researchers report not only the cause of this condition, but also a genetic test that can be applied to breeding programs immediately.
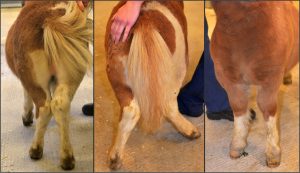
Ponies suffering from skeletal atavism have bowed legs. Photo credit: Ove Wattle.
The researchers first attempted a genome-wide association study, which looks for statistical associations between a disease and variation at a region of the genome, but this approach failed to detect the mutation responsible for SA. To dig a little deeper, they then resequenced the whole genomes of six affected ponies and compared them to the genomes of a pool of control horses—stallions that had at least 50 registered offspring, none of which had SA. They found that the SA-affected ponies suffered from at least one of two partially overlapping deletion mutations, which they labeled Del-1 and Del-2.
These deletions were in sequences unassigned in the Thoroughbred horse reference genome assembly (EquCab2.0), which explains why the genome-wide association study originally failed to find them. These mutations fell in the pseudoautosomal region of the sex chromosomes, a stretch of DNA with a high GC and repeat content that makes it notoriously difficult to sequence and assemble—even in the human genome, about 600 kilobases remain unassembled. The region is very fragmented in the EquCab2.0 genome assembly, and the researchers’ attempts at de novo assembly using short sequence reads from the SA-affected ponies’ genomes also failed.
To circumvent this problem, they used single-molecule real-time (SMRT) sequencing, which enabled them to sequence longer stretches of DNA and pinpoint the deletions to the gene SHOX and the downstream gene CRLF2. Both Del-1 and Del-2 mutants were missing the region downstream of the gene SHOX, which includes the gene CRLF2, but Del-1 mutants were also missing the entire coding region SHOX. Taking into account their functions alone, either gene could plausibly be involved in the deformity. Among the genes’ many roles, CRLF2 is involved in bone metabolism and SHOX is developmentally regulated and activates transcription in cells that develop into bones.
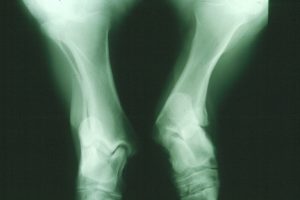
An X-ray image of an atavistic pony’s legs. Photo credit: Göran Dalin.
Although both mutations completely deleted CRLF2, the researchers hypothesize that the effects on SHOX function are the mostly likely cause of SA—whether through deletion of the entire gene or loss of regulatory elements its downstream region. The apparent role of SHOX in humans supports this idea. In humans, haploinsufficiency or deficiency of SHOX is associated with limb deformities, and a 47 kilobase deletion downstream of SHOX is responsible for the skeletal disease Leri-Weill dyschondrosteosis as well as idiopathic short stature. While the results don’t suggest a cure for these human conditions, they did allow the researchers to develop a genetic test for SA in ponies, which—if applied—could allow this crippling condition to be bred out immediately.
CITATION:
Rafati, N.; Andersson, L.; Mikko, S.; Feng, C.; Raudsepp, T.; Pettersson, J.; Janecka, J.; Wattle, O.; Ameur, A.; Thyreen, G.; Eberth, J.; Huddleston, J.; Malig, M.; Bailey, E.; Eichler, E.; Dalin, G.; Chowdary, B.; Anderssson, L.; Lindgren, G.; Rubin, C. Large Deletions at the SHOX Locus in the Pseudoautosomal Region Are Associated with Skeletal Atavism in Shetland Ponies.
G3, 6(7), 2213-2223.
DOI: 10.1534/g3.116.029645
http://www.g3journal.org/content/6/7/2213.full













