The February issue of GENETICS is out now! Check out the highlights below of the full Table of Contents here.
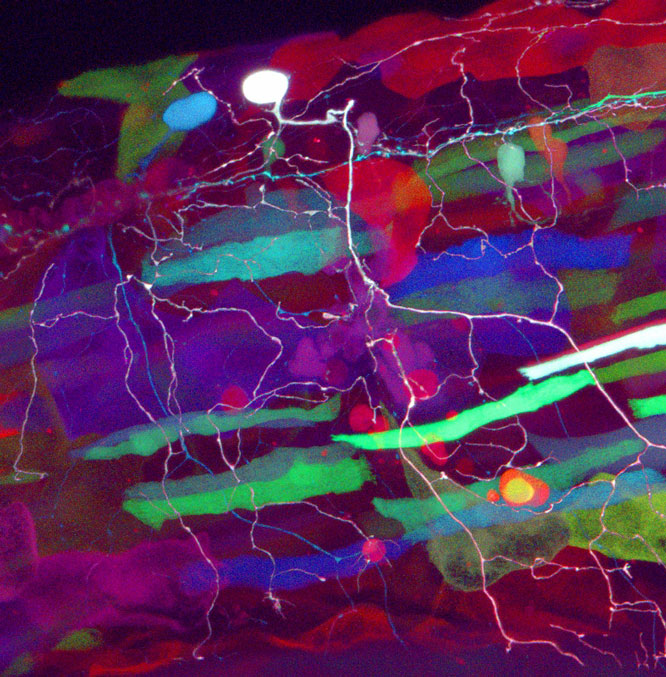
A neuroprotective function of NSF1 sustains autophagy and lysosomal trafficking in Drosophila, pp. 511–522
Daniel T. Babcock, Wei Shen, and Barry Ganetzky
The accumulation of toxic or misfolded proteins is a feature shared by several neurodegenerative diseases. Degradative pathways such as autophagy play a crucial role in eliminating these proteins. Babcock et al. demonstrate a novel neuroprotective role for N-ethyl-maleimide sensitive fusion protein (NSF1) in Drosophila, showing it is required to maintain fusion events required for autophagy and lysosomal trafficking under periods of stress. Expression of NSF1 is able to rescue neurodegeneration in a model of Parkinson’s disease.
Genetic architecture of natural variation of telomere length in Arabidopsis thaliana, pp. 625–635
Nick Fulcher, Astrid Teubenbacher, Envel Kerdaffrec, Ashley Farlow, Magnus Nordborg, and Karel Riha
Telomeres are essential for genome stability because they protect chromosome ends from DNA repair activities. Telomere length varies between species and often between individuals. Telomere length is a highly heritable multigenic trait, but the genes that determine telomere length variation in natural populations are largely unknown. Fulcher et al. examined telomeres in global accessions of Arabidopsis thaliana and used the vast suite of genetic tools available in this species to identify quantitative trait loci underlying telomere length variation.
Marker-based estimation of heritability in immortal populations, pp. 379–398
Willem Kruijer, Martin P. Boer, Marcos Malosetti, Pádraic J. Flood, Bas Engel, Rik Kooke, Joost J. B. Keurentjes, and Fred A. van Eeuwijk
In human genetics, marker-based estimation of heritability assumes only a single individual per genotype can be phenotyped. In contrast, plant traits can be measured on genetically identical replicates, but this advantage is difficult to exploit using most of the current methods. Kruijer et al. propose estimation of heritability using both marker data and replicated phenotypes, as is not uncommon in plant breeding. The proposed estimates are far more accurate than those obtained from genotypic means and classical heritability estimates. Genomic prediction was also improved by this strategy.
Neuropeptide receptors NPR-1 and NPR-2 regulate Caenorhabditis elegans avoidance response to the plant stress hormone methyl salicylate, pp. 523–531
Jintao Luo, Zhaofa Xu, Zhiping Tan, Zhuohua Zhang, and Long Ma
Methyl salicylate (MeSa) is a plant stress hormone that can repel herbivores and attract beneficial insects. Luo et al. found that C. elegans strongly avoids MeSa, and this requires two neuropeptide receptors, NPR-1 at the center of a hub-and-spoke neural circuit and NPR-2 with previously unknown behavioral functions. These receptors act in two distinct interneurons to regulate the avoidance behavior. Similar neuropeptide receptors might regulate other animals’ behavioral responses to MeSa.
A novel nondevelopmental role of the SAX-7/L1CAM cell adhesion molecule in synaptic regulation in Caenorhabditis elegans, pp. 497–509
Karla Opperman, Melinda Moseley-Alldredge, John Yochem, Leslie Bell, Tony Kanayinkal, and Lihsia Chen
The conserved L1CAM family of cell adhesion molecules plays diverse roles essential for nervous system development. Mutations in L1CAMs cause L1 syndrome and are associated with neuropsychiatric and autism spectrum disorders. L1CAM expression in the mature nervous system suggests additional functions besides their well-characterized developmental roles. Opperman et al. used genetic interactions, an inducible L1CAM expression system, and pharmacological reagents to show the C. elegans L1CAM gene, sax-7, plays a previously uncharacterized nondevelopmental role that affects synaptic activity.
The Dnmt1 intrinsically disordered domain regulates genomic methylation during development, pp. 533–541
Ben Shaffer, Serge McGraw, Siyu C. Xiao, Donovan Chan, Jacquetta Trasler, and J. Richard Chaillet
Virtually all sequences in the mammalian genome are modified by DNA methylation, yet the purpose of most methylation is unknown. Shaffer et al. examined the effect of reducing genome methylation in the mouse through mutations in the Dnmt1 cytosine maintenance methyltransferase gene. Homozygous embryos of one mutant line had a profound reduction in global (non-imprinted) methylation, yet maintain imprinted methylation and survived to birth. This work distinguishes two different DNMT1-dependent methylation processes, potentially identifying a much smaller yet functional methylome.
Rapid and precise engineering of the Caenorhabditis elegans genome with lethal mutation co-conversion and inactivation of NHEJ repair, pp. 363–377
Jordan D. Ward
CRISPR/Cas9 is a transformative technology, but efficient recovery of editing events remains a challenge. Here, Ward describes a method in C. elegans to efficiently knock-in epitope tags in 8-9 days using a temperature-sensitive lethal mutation in the pha-1 gene as a co-conversion marker. This selection is stringent, allowing rapid recovery of editing events with minimal screening. Inactivation of non-homologous endjoining further increases knock-in efficiency. Homology arms of 35-80 bp support efficient editing and the DNA break can be at least 54 bp from the insertion site.
A major controversy in codon-anticodon adaptation resolved by a new codon usage index, pp. 573–579
Xuhua Xia
Two alternative hypotheses attribute different benefits to codon-anticodon adaptation. One assumes that protein production is rate limited by both initiation and elongation, and the other states that only initiation efficiency is rate limiting. Here, Xia reanalyzed data from a recent study claiming that elongation efficiency contributes little to protein production in E. coli. Because the original study relied on the codon adaptation index (CAI), the contribution of elongation was significantly underestimated. Xia formulates a new index of translation elongation to facilitate future studies of the translation machinery.
Genetic characterization of human populations: from ABO to a genetic map of the British people, pp. 267–279
Walter Bodmer
The ABO blood groups were the first genetic differences found to vary substantially in frequency between human populations. The discovery of more blood groups, protein variation, and then the HLA tissue typing system eventually provided enough genetic markers to characterize genetic relationships among the major human populations. Now that genetic differences can be detected at the DNA level, an essentially unlimited number of polymorphisms enable increasingly detailed analyses. This Perspectives examines the evolution of methods used to study human polymorphisms and human population relationships, even down to the level of neighboring counties in the UK.